ANOVA Assumptions - R
Contents
ANOVA Assumptions - R#
Shapiro-Wilk Test (on responses)#
Assumption: Normality
Context of Use: t-test, ANOVA, LM, LMM
Reporting: “To test the assumption of conditional normality, a Shapiro-Wilk test was run on the response Y for each combination of levels of factors X1 and X2. All combinations were found to be statistically non-significant except condition (b,b), which showed a statistically significant deviation from normality (W = .794, p < .01).”
# Example data
# df has two factors (X1,X2) each w/two levels (a,b) and continuous response Y
df <- read.csv("data/2F2LBs_normal.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 11.867340 |
| 2 | 2 | a | b | 15.453745 |
| 3 | 3 | b | a | 15.782415 |
| 4 | 4 | b | b | 13.626554 |
| 5 | 5 | a | a | 11.608195 |
| 6 | 6 | a | b | 19.540674 |
| 7 | 7 | b | a | 15.878829 |
| 8 | 8 | b | b | 16.654611 |
| 9 | 9 | a | a | 17.370245 |
| 10 | 10 | a | b | 17.547025 |
| 11 | 11 | b | a | 19.423109 |
| 12 | 12 | b | b | 16.369147 |
| 13 | 13 | a | a | 22.896627 |
| 14 | 14 | a | b | 14.037504 |
| 15 | 15 | b | a | 15.564130 |
| 16 | 16 | b | b | 16.109281 |
| 17 | 17 | a | a | 7.028036 |
| 18 | 18 | a | b | 21.508059 |
| 19 | 19 | b | a | 17.644257 |
| 20 | 20 | b | b | 16.054999 |
## Shapiro-Wilk conditional normality test
# (on the response within each condition)
shapiro.test(df[df$X1 == "a" & df$X2 == "a",]$Y) # condition a,a
shapiro.test(df[df$X1 == "a" & df$X2 == "b",]$Y) # condition a,b
shapiro.test(df[df$X1 == "b" & df$X2 == "a",]$Y) # condition b,a
shapiro.test(df[df$X1 == "b" & df$X2 == "b",]$Y) # condition b,b
Shapiro-Wilk normality test
data: df[df$X1 == "a" & df$X2 == "a", ]$Y
W = 0.97554, p-value = 0.93
Shapiro-Wilk normality test
data: df[df$X1 == "a" & df$X2 == "b", ]$Y
W = 0.93417, p-value = 0.3147
Shapiro-Wilk normality test
data: df[df$X1 == "b" & df$X2 == "a", ]$Y
W = 0.98454, p-value = 0.9914
Shapiro-Wilk normality test
data: df[df$X1 == "b" & df$X2 == "b", ]$Y
W = 0.7938, p-value = 0.003064
Shapiro-Wilk Test (on residuals)#
Assumption: Normality
Context of Use: t-test, ANOVA, LM, LMM
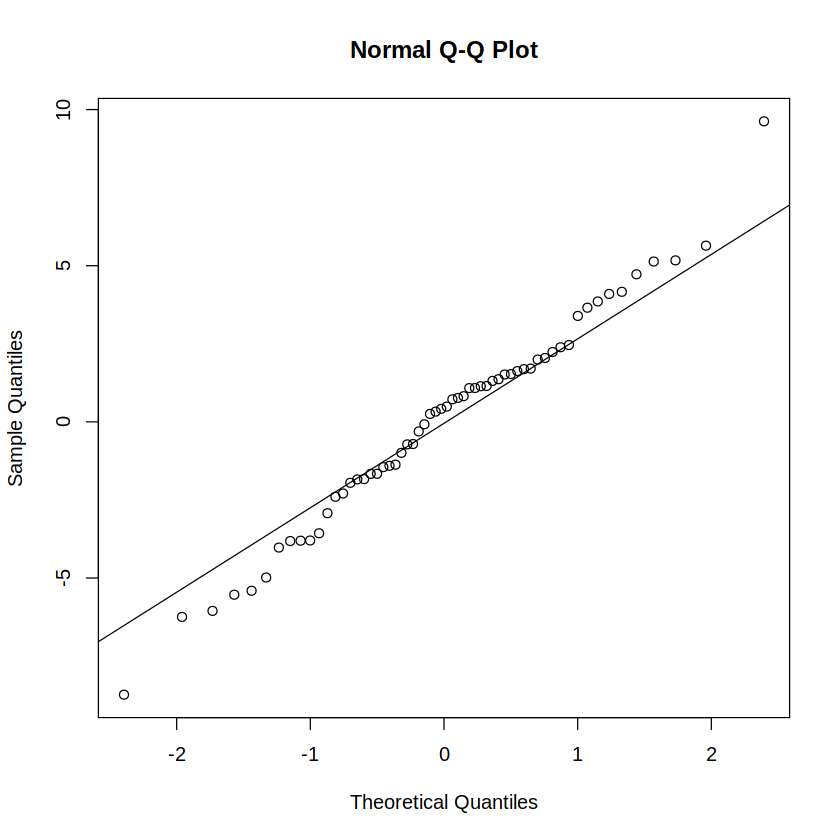
Reporting: “To test the normality assumption, a Shapiro-Wilk test was run on the residuals of a between-subjects full-factorial ANOVA model. The test was statistically non-significant (W = .988, p = .798), indicating compliance with the normality assumption. A Q-Q plot of residuals visually confirms the same (Figure 1).”
# Example data
# df has two factors (X1,X2) each w/two levels (a,b) and continuous response Y
df <- read.csv("data/2F2LBs_normal.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 11.867340 |
| 2 | 2 | a | b | 15.453745 |
| 3 | 3 | b | a | 15.782415 |
| 4 | 4 | b | b | 13.626554 |
| 5 | 5 | a | a | 11.608195 |
| 6 | 6 | a | b | 19.540674 |
| 7 | 7 | b | a | 15.878829 |
| 8 | 8 | b | b | 16.654611 |
| 9 | 9 | a | a | 17.370245 |
| 10 | 10 | a | b | 17.547025 |
| 11 | 11 | b | a | 19.423109 |
| 12 | 12 | b | b | 16.369147 |
| 13 | 13 | a | a | 22.896627 |
| 14 | 14 | a | b | 14.037504 |
| 15 | 15 | b | a | 15.564130 |
| 16 | 16 | b | b | 16.109281 |
| 17 | 17 | a | a | 7.028036 |
| 18 | 18 | a | b | 21.508059 |
| 19 | 19 | b | a | 17.644257 |
| 20 | 20 | b | b | 16.054999 |
m = aov(Y ~ X1*X2, data=df) # make anova model
shapiro.test(residuals(m))
Shapiro-Wilk normality test
data: residuals(m)
W = 0.98751, p-value = 0.7979
par(mfrow=c(1,1))
qqnorm(residuals(m)); qqline(residuals(m)) # Q-Q plot
Anderson-Darling Test (on responses)#
Assumption: Normality
Context of Use: t-test, ANOVA, LM, LMM
Reporting: “To test the assumption of conditional normality, an Anderson-Darling test was run on the response Y for each combination of levels of factors X1 and X2. All combinations were found to be statistically non-significant except condition (b,b), which showed a statistically significant deviation from normality (A = 1.417, p < .001).”
# Example data
# df has two factors (X1,X2) each w/two levels (a,b) and continuous response Y
df <- read.csv("data/2F2LBs_normal.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 11.867340 |
| 2 | 2 | a | b | 15.453745 |
| 3 | 3 | b | a | 15.782415 |
| 4 | 4 | b | b | 13.626554 |
| 5 | 5 | a | a | 11.608195 |
| 6 | 6 | a | b | 19.540674 |
| 7 | 7 | b | a | 15.878829 |
| 8 | 8 | b | b | 16.654611 |
| 9 | 9 | a | a | 17.370245 |
| 10 | 10 | a | b | 17.547025 |
| 11 | 11 | b | a | 19.423109 |
| 12 | 12 | b | b | 16.369147 |
| 13 | 13 | a | a | 22.896627 |
| 14 | 14 | a | b | 14.037504 |
| 15 | 15 | b | a | 15.564130 |
| 16 | 16 | b | b | 16.109281 |
| 17 | 17 | a | a | 7.028036 |
| 18 | 18 | a | b | 21.508059 |
| 19 | 19 | b | a | 17.644257 |
| 20 | 20 | b | b | 16.054999 |
library(nortest)
ad.test(df[df$X1 == "a" & df$X2 == "a",]$Y) # condition a,a
ad.test(df[df$X1 == "a" & df$X2 == "b",]$Y) # condition a,b
ad.test(df[df$X1 == "b" & df$X2 == "a",]$Y) # condition b,a
ad.test(df[df$X1 == "b" & df$X2 == "b",]$Y) # condition b,b
Anderson-Darling normality test
data: df[df$X1 == "a" & df$X2 == "a", ]$Y
A = 0.23266, p-value = 0.7557
Anderson-Darling normality test
data: df[df$X1 == "a" & df$X2 == "b", ]$Y
A = 0.35901, p-value = 0.4023
Anderson-Darling normality test
data: df[df$X1 == "b" & df$X2 == "a", ]$Y
A = 0.16003, p-value = 0.934
Anderson-Darling normality test
data: df[df$X1 == "b" & df$X2 == "b", ]$Y
A = 1.4167, p-value = 0.0007191
Anderson-Darling Test (on residuals)#
Assumption: Normality
Context of Use: t-test, ANOVA, LM, LMM
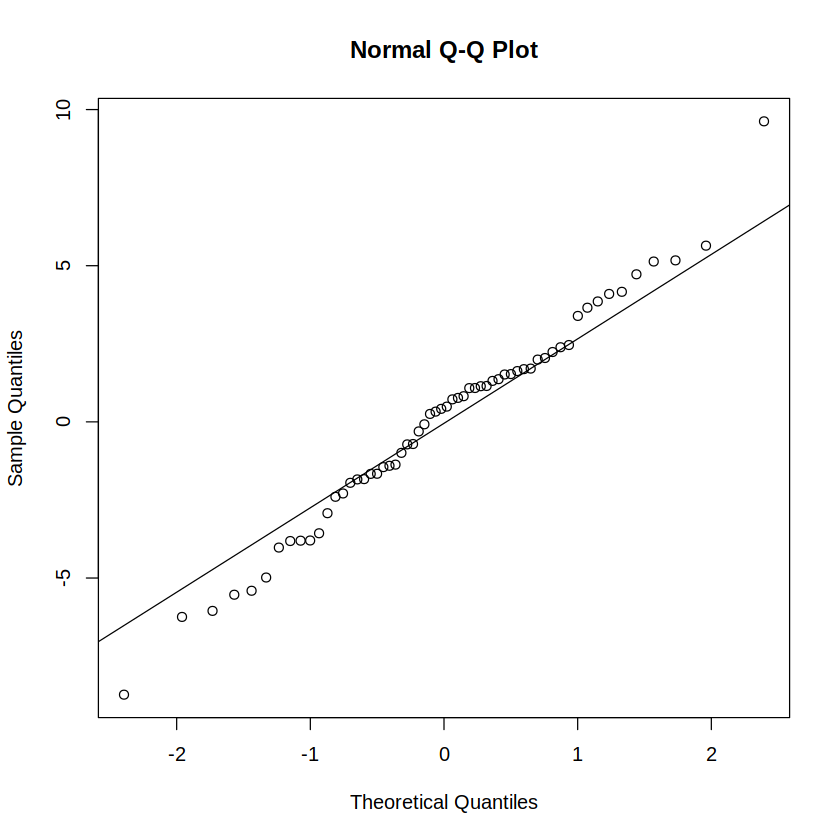
Reporting: “To test the normality assumption, an Anderson-Darling test was run on the residuals of a between-subjects full-factorial ANOVA model. The test was statistically non-significant (A = 0.329, p = .510), indicating compliance with the normality assumption. A Q-Q plot of residuals visually confirms the same (Figure 1).”
# Example data
# df has two factors (X1,X2) each w/two levels (a,b) and continuous response Y
df <- read.csv("data/2F2LBs_normal.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 11.867340 |
| 2 | 2 | a | b | 15.453745 |
| 3 | 3 | b | a | 15.782415 |
| 4 | 4 | b | b | 13.626554 |
| 5 | 5 | a | a | 11.608195 |
| 6 | 6 | a | b | 19.540674 |
| 7 | 7 | b | a | 15.878829 |
| 8 | 8 | b | b | 16.654611 |
| 9 | 9 | a | a | 17.370245 |
| 10 | 10 | a | b | 17.547025 |
| 11 | 11 | b | a | 19.423109 |
| 12 | 12 | b | b | 16.369147 |
| 13 | 13 | a | a | 22.896627 |
| 14 | 14 | a | b | 14.037504 |
| 15 | 15 | b | a | 15.564130 |
| 16 | 16 | b | b | 16.109281 |
| 17 | 17 | a | a | 7.028036 |
| 18 | 18 | a | b | 21.508059 |
| 19 | 19 | b | a | 17.644257 |
| 20 | 20 | b | b | 16.054999 |
library(nortest)
m = aov(Y ~ X1*X2, data=df) # make anova model
ad.test(residuals(m))
Anderson-Darling normality test
data: residuals(m)
A = 0.32859, p-value = 0.5102
par(mfrow=c(1,1))
qqnorm(residuals(m)); qqline(residuals(m)) # Q-Q plot
Levene’s Test#
Assumption: Homoscedasticity (Homogeneity of Variance)
Context of Use: t-test, ANOVA, LM, LMM
Reporting: “To test the homoscedasticity assumption, Levene’s test was run on a between-subjects full-factorial ANOVA model. The test was statistically significant (F(3, 56) = 3.97, p < .05), indicating a departure from homoscedasticity.”
# Example data
# df has two factors (X1,X2) each w/two levels (a,b) and continuous response Y
df <- read.csv("data/2F2LBs_normal.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 11.867340 |
| 2 | 2 | a | b | 15.453745 |
| 3 | 3 | b | a | 15.782415 |
| 4 | 4 | b | b | 13.626554 |
| 5 | 5 | a | a | 11.608195 |
| 6 | 6 | a | b | 19.540674 |
| 7 | 7 | b | a | 15.878829 |
| 8 | 8 | b | b | 16.654611 |
| 9 | 9 | a | a | 17.370245 |
| 10 | 10 | a | b | 17.547025 |
| 11 | 11 | b | a | 19.423109 |
| 12 | 12 | b | b | 16.369147 |
| 13 | 13 | a | a | 22.896627 |
| 14 | 14 | a | b | 14.037504 |
| 15 | 15 | b | a | 15.564130 |
| 16 | 16 | b | b | 16.109281 |
| 17 | 17 | a | a | 7.028036 |
| 18 | 18 | a | b | 21.508059 |
| 19 | 19 | b | a | 17.644257 |
| 20 | 20 | b | b | 16.054999 |
library(car) # for leveneTest and Anova
leveneTest(Y ~ X1*X2, data=df, center=mean)
# if a violation occurs and only a t-test is needed, use a Welch t-test
t.test(Y ~ X1, data=df, var.equal=FALSE) # Welch t-test
# if a violation occurs and an ANOVA is needed, use a White-adjusted ANOVA
m = aov(Y ~ X1*X2, data=df)
Anova(m, type=3, white.adjust=TRUE)
Loading required package: carData
| Df | F value | Pr(>F) | |
|---|---|---|---|
| <int> | <dbl> | <dbl> | |
| group | 3 | 3.965124 | 0.01238959 |
| 56 | NA | NA |
Welch Two Sample t-test
data: Y by X1
t = 0.80702, df = 43.125, p-value = 0.4241
alternative hypothesis: true difference in means between group a and group b is not equal to 0
95 percent confidence interval:
-1.188936 2.775540
sample estimates:
mean in group a mean in group b
15.56348 14.77018
Coefficient covariances computed by hccm()
| Df | F | Pr(>F) | |
|---|---|---|---|
| <dbl> | <dbl> | <dbl> | |
| (Intercept) | 1 | 103.0349856 | 2.652463e-14 |
| X1 | 1 | 0.4129862 | 5.230802e-01 |
| X2 | 1 | 8.0591851 | 6.296776e-03 |
| X1:X2 | 1 | 3.6441353 | 6.139637e-02 |
| Residuals | 56 | NA | NA |
Brown-Forsythe Test#
Assumption: Homoscedasticity (Homogeneity of Variance)
Context of Use: t-test, ANOVA, LM, LMM
Reporting: “To test the homoscedasticity assumption, the Brown-Forsythe test was run on a between-subjects full-factorial ANOVA model. The test was statistically significant (F(3, 56) = 3.75, p < .05), indicating a departure from homoscedasticity.”
# Example data
# df has two factors (X1,X2) each w/two levels (a,b) and continuous response Y
df <- read.csv("data/2F2LBs_normal.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 11.867340 |
| 2 | 2 | a | b | 15.453745 |
| 3 | 3 | b | a | 15.782415 |
| 4 | 4 | b | b | 13.626554 |
| 5 | 5 | a | a | 11.608195 |
| 6 | 6 | a | b | 19.540674 |
| 7 | 7 | b | a | 15.878829 |
| 8 | 8 | b | b | 16.654611 |
| 9 | 9 | a | a | 17.370245 |
| 10 | 10 | a | b | 17.547025 |
| 11 | 11 | b | a | 19.423109 |
| 12 | 12 | b | b | 16.369147 |
| 13 | 13 | a | a | 22.896627 |
| 14 | 14 | a | b | 14.037504 |
| 15 | 15 | b | a | 15.564130 |
| 16 | 16 | b | b | 16.109281 |
| 17 | 17 | a | a | 7.028036 |
| 18 | 18 | a | b | 21.508059 |
| 19 | 19 | b | a | 17.644257 |
| 20 | 20 | b | b | 16.054999 |
library(car) # for leveneTest, Anova
leveneTest(Y ~ X1*X2, data=df, center=median)
# if a violation occurs and only a t-test is needed, use a Welch t-test
t.test(Y ~ X1, data=df, var.equal=FALSE) # Welch t-test
# if a violation occurs and an ANOVA is needed, use a White-adjusted ANOVA
m = aov(Y ~ X1*X2, data=df)
Anova(m, type=3, white.adjust=TRUE)
| Df | F value | Pr(>F) | |
|---|---|---|---|
| <int> | <dbl> | <dbl> | |
| group | 3 | 3.750252 | 0.01587369 |
| 56 | NA | NA |
Welch Two Sample t-test
data: Y by X1
t = 0.80702, df = 43.125, p-value = 0.4241
alternative hypothesis: true difference in means between group a and group b is not equal to 0
95 percent confidence interval:
-1.188936 2.775540
sample estimates:
mean in group a mean in group b
15.56348 14.77018
Coefficient covariances computed by hccm()
| Df | F | Pr(>F) | |
|---|---|---|---|
| <dbl> | <dbl> | <dbl> | |
| (Intercept) | 1 | 103.0349856 | 2.652463e-14 |
| X1 | 1 | 0.4129862 | 5.230802e-01 |
| X2 | 1 | 8.0591851 | 6.296776e-03 |
| X1:X2 | 1 | 3.6441353 | 6.139637e-02 |
| Residuals | 56 | NA | NA |
Mauchly’s Test of Sphericity#
Assumption: Sphericity
Context of Use: repeated measures ANOVA
Reporting: “To test the sphericity assumption for repeated measures ANOVA, Mauchly’s test of sphericity was run on a mixed factorial ANOVA model with a between-subjects factor X1 and a within-subjects factor X2. The test was statistically significant for both X2 (W = .637, p < .01) and X1×X2 (W = .637, p < .01), indicating sphericity violations. Accordingly, the Greenhouse-Geisser correction was used when reporting these ANOVA results.”
# Example data
# df has subjects (S), one between-Ss factor (X1), and one within-Ss factor (X2)
df <- read.csv("data/2F23LMs_mauchly.csv")
head(df, 20)
| S | X1 | X2 | Y | |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <dbl> | |
| 1 | 1 | a | a | 20.2145 |
| 2 | 1 | a | b | 23.7485 |
| 3 | 1 | a | c | 20.7960 |
| 4 | 2 | a | a | 20.8805 |
| 5 | 2 | a | b | 23.2595 |
| 6 | 2 | a | c | 19.1305 |
| 7 | 3 | a | a | 21.2635 |
| 8 | 3 | a | b | 23.4945 |
| 9 | 3 | a | c | 20.8545 |
| 10 | 4 | a | a | 20.7080 |
| 11 | 4 | a | b | 23.9220 |
| 12 | 4 | a | c | 18.2575 |
| 13 | 5 | a | a | 21.0075 |
| 14 | 5 | a | b | 23.4700 |
| 15 | 5 | a | c | 17.7105 |
| 16 | 6 | a | a | 19.9115 |
| 17 | 6 | a | b | 24.2975 |
| 18 | 6 | a | c | 19.8550 |
| 19 | 7 | a | a | 22.1050 |
| 20 | 7 | a | b | 25.3800 |
library(ez) # for ezANOVA
df <- read.csv("data/2F23LMs_mauchly.csv")
df$S = factor(df$S) # Subject id is nominal
m = ezANOVA(dv=Y, between=c(X1), within=c(X2), wid=S, type=3, data=df) # use c() for >1 factors
m$Mauchly # p<.05 indicates a sphericity violation for within-Ss effects
Warning message:
“Converting "X2" to factor for ANOVA.”
Warning message:
“Converting "X1" to factor for ANOVA.”
| Effect | W | p | p<.05 | |
|---|---|---|---|---|
| <chr> | <dbl> | <dbl> | <chr> | |
| 3 | X2 | 0.6370236 | 0.008782794 | * |
| 4 | X1:X2 | 0.6370236 | 0.008782794 | * |

